An update and expansion on “The Water Professional’s Guide to COVID-19”
Rasha Maal-Bared, Naoko Munakata, Kyle Bibby, Kari Brisolara, Charles Gerba, Mark Sobsey, Scott Schaefer, Jay Swift, Lee Gary, Samendra Sherchan, Akin Babatola, Lola Olabode, Robert Reimers, Robert Bastian, and Albert Rubin
Primary Messages
The research community has yet to find evidence of survival of infective COVID-19 virus in feces and in wastewater systems. Efforts are ongoing to assess the relative risk of exposure to wastewater.
Current efforts estimate numbers of infections in the community and support public health surveillance by detecting the virus in wastewater using molecular techniques that identify presence and quantities of genetic material (RNA). This method does not assess virus viability or infectivity. Future research on COVID-19 virus is encouraged to investigate concentrations of infective virus in feces (if any), shedding patterns in patients and factors influencing persistence of infective virus in plumbing and collection systems
Research is ongoing, but experts currently believe that exposure to wastewater is not a significant transmission route for COVID-19 virus.
WEF continues to work on improving protection of wastewater workers from pathogen exposure — especially via aerosols — by forming a Blue Ribbon Panel to address the issue of required personal protective equipment, as well as protective work and hygiene practices.
Preamble
On February 19, 2020, WEF published The Water Professional’s Guide to COVID-19. The Guide was meant to increase water sector awareness of COVID-19 virus and any water and wastewater-related issues and relevant resources. The content was reprinted in the April 2020 issue of Water Environment & Technology. Since then, the World Health Organization (WHO) has declared COVID-19 a pandemic and the disease has spread to more than 210 countries with more research on the virus being published every day.
In an effort to keep the water community informed of COVID-19 developments, the Waterborne Infectious Disease Outbreak Control (WIDOC) working group has prepared an update to highlight the latest scientific findings, as well as topics not previously addressed. The goal is to contextualize these new results and state the implications and significance from a water and wastewater collection and treatment, public health and water resource recovery facility (WRRF) worker perspective.
This update reinforces our previous recommendations, which align with the U.S. Centers for Disease Control and Prevention (CDC), the U.S. Occupational Safety and Health Administration (OSHA), WHO and the Pan American Health Organization (PAHO).
There is still no evidence that coronavirus survives the disinfection processes utilized for drinking water and wastewater treatment. No coronavirus-specific protections are recommended for workers working in wastewater treatment and collection systems. However, as data emerges almost daily with regard to this virus, heightened vigilance in compliance with existing personal protective procedures is appropriate to control exposure.
Table of Contents
- Preamble
- Does detecting COVID-19 virus RNA mean the virus is infective?
- COVID-19 virus nomenclature
- Can COVID-19 virus spread through the sewerage system?
- Can COVID-19 virus spread through the fecal-oral contamination route?
- Can COVID-19 virus be transmitted via aerosols?
- Evidence of COVID-19 virus survival on surfaces
- Comparing SARS, MERS and COVID-19 viruses
- Authors
Does detecting COVID-19 virus RNA mean the virus is infective?
No. As explained below, viruses can be detected using a variety of methods. Some test whether the virus can infect cells, while others just look for nucleic acids (also called RNA or RNA fragments). But viruses need much more than just their RNA to successfully infect cells, so detecting RNA is a little bit like detecting an antibody — it tells you that the virus was once there, but not whether it is still there and is infectious now.
Virus structure and function
Not all pathogenic viruses detected in the built environment by molecular methods are infectious (Prussin et al. 2020). To understand the difference between detecting RNA and detecting infective virus, we must first understand the basic structure and biology of viruses.
When outside a host, viruses are considered non-living or inert particles called virions. Virions are not cells. They are made up of nucleic acids — DNA or RNA — contained in a protein coat (called a capsid) and in some cases, such as COVID-19 virus, they are surrounded by a lipid envelope. Most importantly, they do not have the necessary parts to replicate (i.e. produce copies of themselves) on their own, so they need to find a host cell, hijack it, and make it produce copies of the virus. Figure 1 depicts COVID-19 virus attaching to a receptor on a host cell.
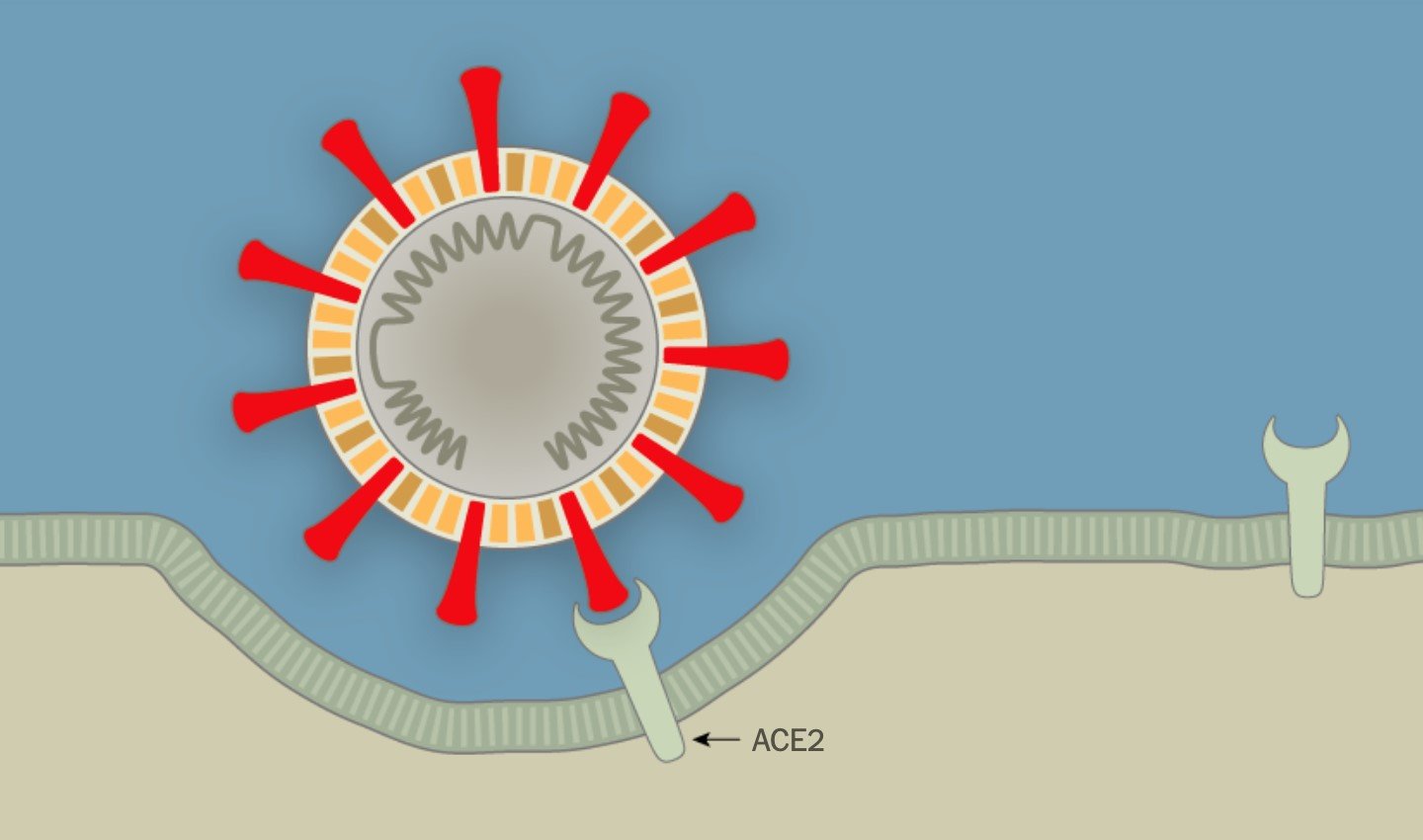
Figure 1. COVID virus attaching to host cells (New York Times Image, 2020).
In this process, the red spikes on the surface of the COVID-19 virus (called the S proteins) are extremely important. These S proteins allow the virus to attach to the host cell receptors (ACE2) and gain entry into the host cell to replicate. This means that, in principle, all cells carrying ACE2 receptors in the respiratory or intestinal tract can potentially be hijacked by COVID-19 virus.
What methods do we use to detect viruses and what can they tell us about risk?
Environmental virology has undergone many changes in detection methods over the last century. Identification procedures have evolved from methods relying on production of disease in laboratory animals, to detection of detrimental changes in cell cultures, to immunologic approaches, to detection of the viral genome or nucleic acids (Metcalf et al., 1995) and most recently the addition of transfection assays.
The advent of cell culture assays was a major catalyst for improving our understanding of waterborne viruses and their transmission routes. In cell culture assays, a water or wastewater sample is added to a prepared population of suitable host cells (containing the right receptors for the virus under investigation) in a petri-dish, flask, or other container to examine virus presence and infectivity in the sample by monitoring host cell changes. When the virus in the sample invades the host cells and uses them to replicate, eventually the host cell will display visible negative effects. These effects on cells are called cytopathic effects (CPE) and can take on several forms, the most extreme being host cells bursting (or lysing) creating a hole or “plaque” in the smooth lawn of host cells. These plaques are then counted and the concentration is reported as plaque forming units (PFU) per volume. This is similar to bacteria tests, where bacteria are grown on a plate of suitable nutrient media, and resulting colonies are counted and reported as colony forming units per volume. Alternatively, virus infection of host cells can be detected and quantified as a most probable number (MPN) concentration, which is based on the number of virus CPE-positive and virus CPE-negative host cell culture plates or tubes. The examination of the cell cultures for such CPE is often done using a microscope. This MPN method is essentially the same as those used for bacteria quantification by a multiple fermentation tube method or a quantitative well tray method.
While cell culture assays give you a clear idea of risk or infectivity, researchers often opt for other detection methods for several reasons. Infective cell cultures are very difficult to perform, with some viruses displaying very slow growth to no visible growth at all. Cell culture assays try to replicate the perfect chemical, biological, and environmental conditions that result in host cell infection. In the case of emerging viruses, such as the COVID-19 virus, we may not even fully understand these conditions yet. Cell culture assays are expensive, time-consuming, susceptible to contamination, and require specialized equipment and reagents. Some viruses result in changes to host cells that are not easily seen or documented. In some cases, biosafety regulations prohibit researchers from handling or isolating some viable or infective viruses unless their laboratories have a certain level of biosafety certification (i.e., protections in place to ensure the virus does not harm lab staff, public health, or the environment).
Biosafety levels (BSL) range from Level 1 to Level 4, with BSL4 requiring the highest levels of precautions. Most academic labs have a Biosafety Level 2 (BSL2) certification or license, which allows the collection, import, concentration, and handling of organisms such as enteroviruses, Adenovirus, Salmonella, Campylobacter, Cryptosporidium, Giardia, Pseudomonas, etc. Many emerging viruses — such as COVID-19 virus — require a BSL3 lab. Handling Ebolavirus requires a BSL4 laboratory.
In the 1980s, researchers started using nucleic acid amplification techniques or molecular techniques, such as polymerase chain reaction (PCR), to detect unique, microorganism-specific nucleic acid sequences in clinical and environmental samples.
PCR is a technique that amplifies (i.e., synthesizes many copies) of a specific DNA segment of interest by using short, synthetic nucleotide sequences called primers that bind to unique regions of the target genome enabling specific identification of the organism. To accomplish this, researchers collect a sample, concentrate it through filtration and elution (i.e., detaching particles from filters after concentration) or other physical and chemical methods (such as centrifugation and chemical precipitation), and, then, chemically disrupt the virus capsids to access the RNA, a process called RNA extraction and purification. Reverse transcriptase-PCR (RT-PCR) is a type of PCR used to detect the presence of RNA viruses. Since PCR is designed to work on DNA, RT-PCR first requires producing a complementary DNA (cDNA) strand from the small amount of viral RNA in the sample before amplification by PCR. Ultimately, researchers combine the nucleic acids from their sample with primers, reagents, and fluorescent dyes that will only fluoresce if the primers attach to their target sequence and initiate the nucleic acid amplification (copying) process. The intensity of the fluorescent signal produced corresponds to the amount of target RNA in the sample analyzed — but does not measure virus infectivity or viability. With the use of positive controls with known target virus concentrations, the intensity of the fluorescence can be used for quantification purposes.
While much simpler than cell culture, molecular methods also present many challenges. Methodologically, the degree of sensitivity and specificity achieved by RT-PCR depend on both the effectiveness of the recovery of virus from samples through concentration and purification processes and the degree of final purity of recovered virus through removal of PCR-inhibiting substances.
Another challenge with molecular methods is the translation of nucleic acid testing results to public health risk. Results of nucleic acid-based testing are an expression of total virus genetic material present, with no distinction between infectious and non-infectious particles (Metcalf et al., 1995).
PCR cannot measure to what degree a virus is affected by different disinfectants. This is because PCR targets specific parts or fragments of the RNA and does not take into account the type and severity of damage caused to the genome by disinfectants (e.g., RNA structural modification, phosphate backbone scission, etc.) (Torrey et al., 2019). Mainly, a negative cell culture result (i.e., loss of infectivity) is not always associated with a negative PCR result. However, a negative PCR result usually means loss of all activity and removal of risk. Ultimately, a PCR positive result does not provide evidence for public health risk from infectious virus (Metcalf et al., 1995) or treatment failure (Torrey et al., 2019) and is, therefore, not independently a cause for action by utilities or regulatory agencies.
More direct approaches have been developed to evaluate outstanding risk from viruses after exposure to stress, treatments, or disinfection. Some researchers assess infectivity by utilizing novel transfection methods, where a virus genome can be inserted directly into host cells to observe their ability to create viral progeny after disinfection (Torrey et al., 2019). Other researchers have introduced an initial cell culture step prior to running a PCR so that infective viruses predominate in the sample. Unfortunately, studies attempting to establish known relationships between infectious, cultivable virus assays (PFU) and nucleic acid-based tests (PCR) are ambiguous and contradictory in the ability to establish the concentration ratios between the two metrics (RNA and infectivity).
Recent efforts to monitor COVID-19 virus in wastewater in the Netherlands, Australia and the United States have relied on RT-PCR as a detection tool. The main objective of these studies was to support public health surveillance and estimate the number of infections in the community (Mallapati, 2020).
This is not a new approach. Environmental virologists realized the value of poliovirus surveillance by cell culture assays in wastewater in the 1940s. They observed that when symptomatic cases in the community were prevalent, polioviruses more frequently detected and at higher concentrations in wastewater. This approach enabled estimating the extent of infection and the ratio of symptomatic to asymptomatic infections in the community, based on shedding estimates from individual patients and what was detected in community wastewater (Metcalf et al., 1995). Since then, wastewater epidemiology approaches have also been used effectively for early detection of disease outbreaks and informing the efficacy of public health interventions for enteric viruses, such as polio eradication by polio immunization with vaccines and norovirus and hepatitis A virus control measures (Ahmad et al., 2020). See Figure 2 for more detail on COVID-19 virus detection methods.
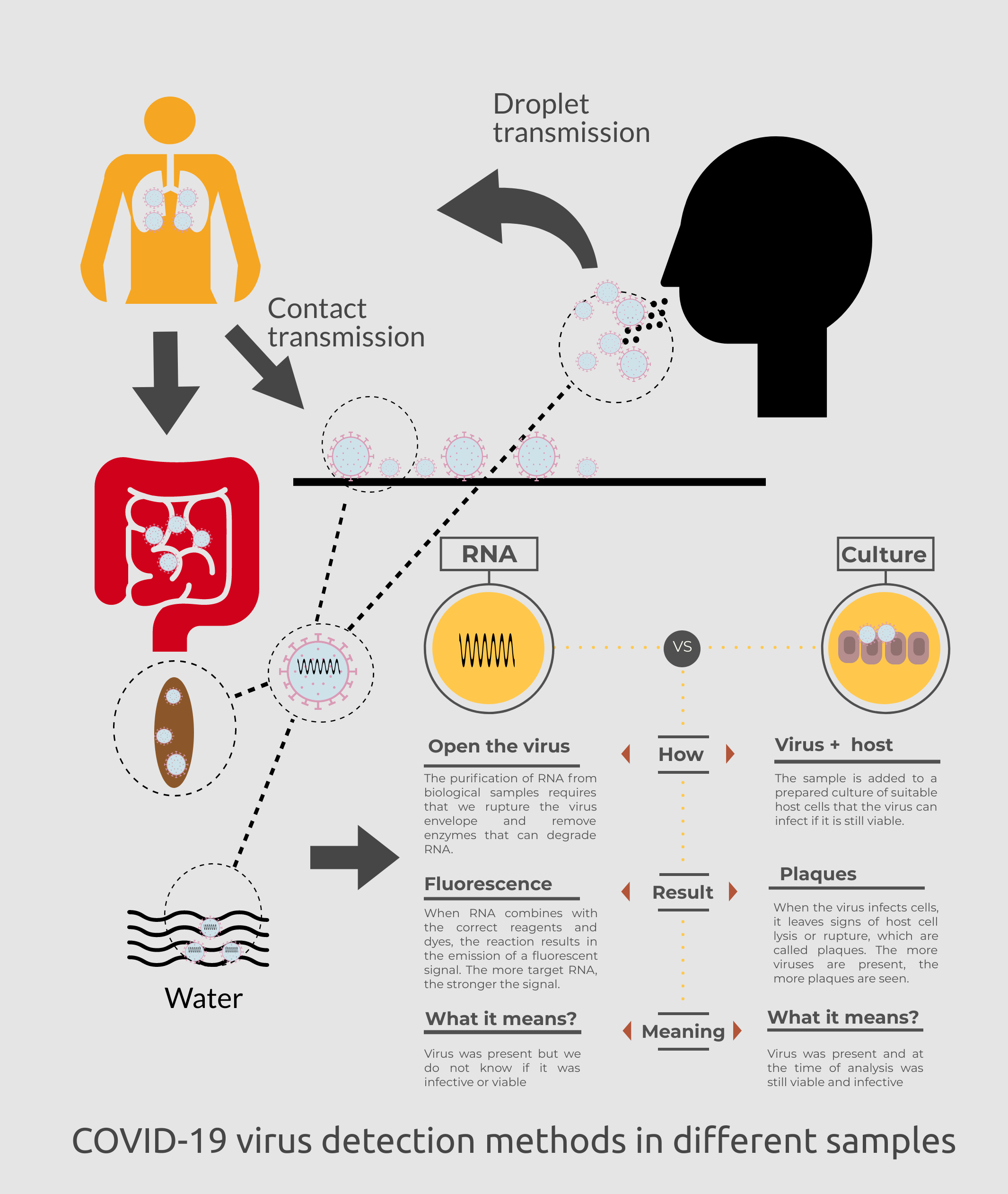
Figure 2. COVID-19 virus potential detection methods for clinical and environmental samples. (Get PDF)
How does environmental stress and disinfection impact virus infectivity?
Human pathogenic viruses enter the environment through feces, urine, and other bodily secretions, including saliva, sputum, blood, and sweat (Rodriguez-Lazaro et al., 2012). As a result, water, wastewater, wastewater sludge, air, surfaces, produce, shellfish, and animal products can be sources of environmental exposure to human pathogenic viruses. Virus survival in these matrices is a function of the virus properties and the matrix, but virus infectivity is a more complicated matter as shown in Figure 3.
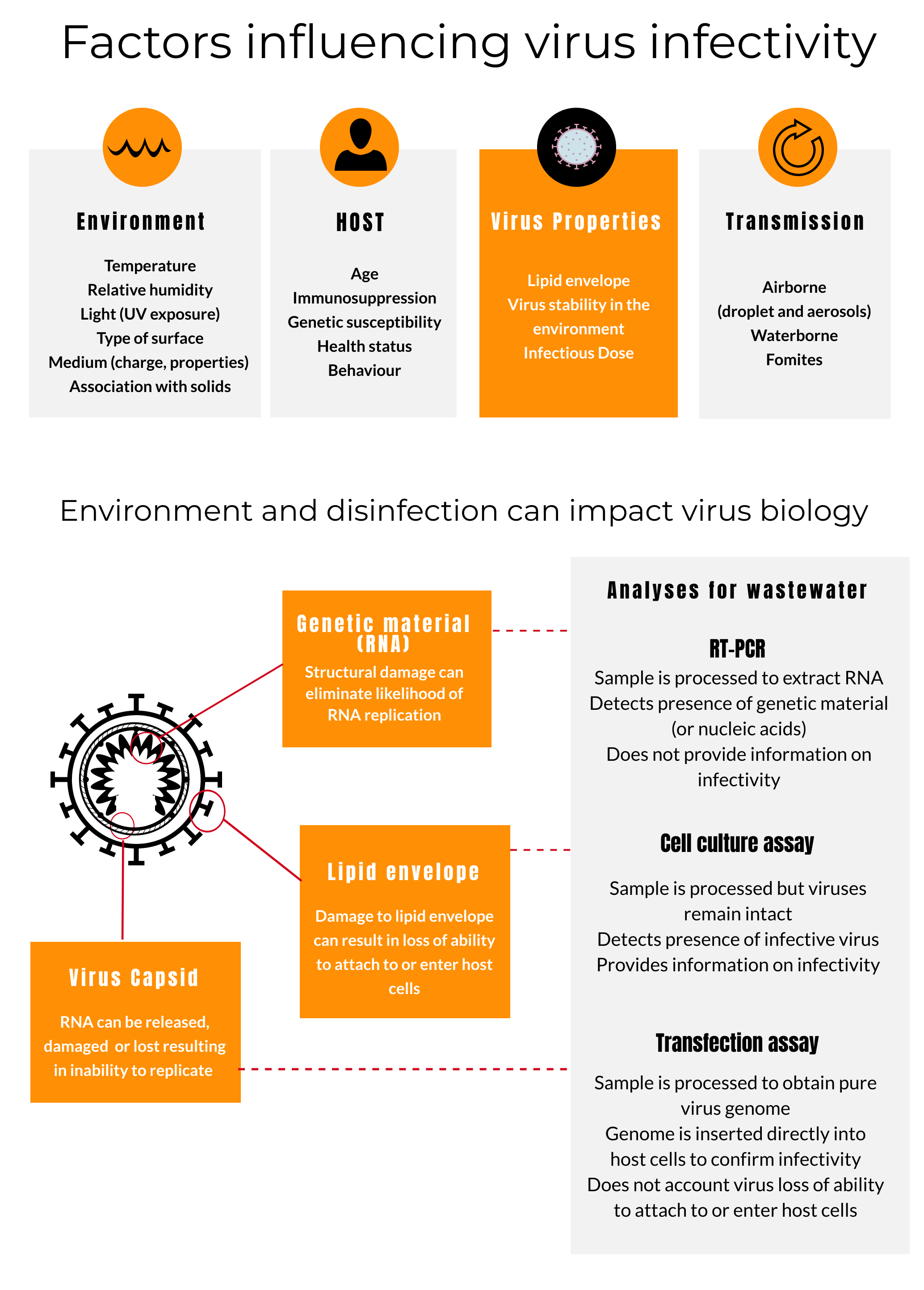
Figure 3. Factors influencing virus infectivity. (Get PDF)
Infectivity is the capacity of a virus to attach to a host cell, inject its nucleic acids into the host cell, and then replicate following its earlier release from an infected individual (Wigginton et al., 2012) at high enough concentrations and for enough time to cause infection in a susceptible individual. Infectivity is highly dependent on properties of the virus, the host, environmental conditions, and the mode of transmission (Prussin et al., 2020; Rodriguez-Lazaro et al., 2012). To determine infectivity, researchers use cell culture assays or transfection assays. See Figure 3 for more detail.
Disinfection strategies for wastewater or surfaces contaminated with COVID-19 virus can target one or more virus structures involved with any of these three functions: the genomic material (RNA for COVID-19 virus), the protein capsid, or the lipid envelope. Although some disinfectants can alter viral nucleic acids, many simply alter the lipid envelope or the viral capsid that contains the nucleic acids; others, such as strong oxidants like free chlorine and ozone alter multiple of these virus targets (Morato et al., 2003). In the case of COVID-19-virus, disrupting the lipid envelop can eliminate risk since the virus can no longer attach to and enter host cells. This is why enveloped viruses are considered amongst the most susceptible organisms to disinfection.
Depending on which virus structures are targeted, efficacy of disinfection and resulting viral infectivity can be altered to different degrees. For example, Wigginton et al. (2012) found that exposing MS2 bacteriophage to ultraviolet (UV) light both damages the viral genome, so that it can no longer replicate itself in the host, and breaks down its capsid inhibiting injection of the genomic material into the host cell. Chlorine attacks the genome, prevents it from replicating, and destroys its injection function (Wigginton et al., 2012). RNA testing does not tell us if nucleic acids have been sufficiently damaged and the virus is no longer infectious. However, viruses rendered non-infectious can often still be detected by amplifying their nucleic acids.
COVID-19 Virus Nomenclature
Summary: On January, 12, 2020, WHO announced that a novel human coronavirus had been identified in samples obtained from viral pneumonia cases from Wuhan, China, and that initial analysis of virus genetic sequences suggested that this was the cause of the outbreak. This new virus is referred to as SARS-CoV-2, and the associated disease is COVID-19. The name of the virus, however, was changed frequently and appeared in the literature as 2019-nCoV, SARS-CoV-2, HCoV-19, COVID-19 virus, and the virus responsible for COVID-19.
Implications for water professionals: In official communications, it is recommended to use COVID-19 when referring to the novel coronavirus disease. When referring to the virus itself, there is less specific guidance due to variations in published literature. Many water organizations use COVID-19 virus or the virus resulting in COVID-19 to avoid confusion.
Details: On January, 12, 2020, WHO determined that a novel human coronavirus that had been identified in samples obtained from viral pneumonia cases from Wuhan, China, was the cause of an outbreak. The virus was temporarily referred to as 2019-nCoV by the scientific community. On February, 11, 2020, WHO named the coronavirus-associated acute respiratory disease as coronavirus disease 19 (COVID-19). Following SARS-CoV-1 and MERS-CoV, this was the third documented spillover of an animal coronavirus to humans in only two decades.
The Coronaviridae Study Group (CSG) of the International Committee on Taxonomy of Viruses responsible for developing the classification of viruses of the family Coronaviridae suggested designating the virus as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) on the basis of a phylogenetic analysis of related coronaviruses.
The distinction between the virus and disease is important; similar to saying the variola virus causes smallpox and HIV causes AIDS, SARS-CoV-2 causes COVID-19.
CGS had decided that the SARS-CoV-2 belongs to the same species of coronavirus that caused an outbreak of severe acute respiratory syndrome (SARS) in 2002 and 2003 and is a variant (or sister clade) of SARS-CoV-1. By choosing this name, CGS created some confusion as the new name incorrectly implies to those with little virology literacy that this novel virus is epidemiologically or clinically similar SARS-CoV-1 or its resulting disease; it is not.
COVID-19 virus is different from all other SARS-like or SARS-related coronaviruses, which are characterised mainly by their genome sequence. The new name is also inconsistent with the disease name COVID-19. Other names have been proposed, such as human coronavirus 2019 (HCoV-19), to avoid confusion and facilitate communication.
Ultimately, several global health organizations feared that linking the novel coronavirus to SARS would have unintended consequences and confuse the public from a risk communication perspective. As a result WHO, the Water Research Foundation, and other relevant organizations have referred to the coronavirus responsible for the current pandemic as 'the virus responsible for COVID-19' or 'the COVID-19 virus'. Neither of these designations will replace the scientific designation of the virus that the CGS study group has chosen.
Can COVID-19 virus spread through the sewerage system?
Summary: Despite prolonged shedding of the virus RNA in feces, experts believe that exposure to wastewater is not a significant transmission route for COVID-19. However, most scientific efforts related to wastewater thus far have not looked for infective viruses. Instead they have focused on quantifying virus RNA levels to try to estimate the infection rate in the population, thereby aiding public health surveillance and infection control in the population. It is important to note that detection of virus RNA by RT-PCR provides no indication that the virus is infectious.
Implications for water professionals: While much is still unknown about COVID-19 virus shedding and transmission, CDC and WHO state that current evidence does not support that COVID-19 virus is transmitted via wastewater.
Water professionals should be able to communicate that the detection of COVID-19 virus RNA in wastewater does not translate to public health risk and does not result in the need for change in operations or procedures by utilities. The preliminary data from treated effluent suggests that wastewater treatment processes that are in place already are sufficient for COVID-19 virus control. This data corresponds with CDC, OSHA, WHO, and WEF recommendations. The data produced from surveillance of RNA could indicate the scale of the outbreak and aid in emergency response planning in communities and at utilities (i.e., prediction of absenteeism and critical staff management), but these data need to be interpreted with caution.
Details: At the time of publication of this update, three relevant studies have investigated the use of wastewater-based epidemiology approaches to support public health surveillance for COVID-19 infections in communities. One publication is a pre-proof from Australia by Ahmed et al. (2020), while the other two are pre-prints (i.e., have not been peer-reviewed yet) by Medema et al. (2020) from KWR Watercycle Research Institute in the Netherlands and by Wu et al. (2020b) from Biobot in Massachusetts. Wastewater-based epidemiology relies on using influent COVID-19 virus RNA concentrations in relation to per capita mass loads, daily flows, and size of WRRF population served to estimate the number of infections on a population-scale within sewerage watershed boundaries.
The work from Australia was the first peer-reviewed publication to confirm the usefulness of the wastewater-based epidemiology approach for tracking COVID-19 infections in communities. They tested COVID-19 virus RNA concentrations in the wastewater and used these concentrations to estimate the number of infected individuals in the catchment via Monte Carlo simulations. Given the uncertainty and variation in the input parameters (e.g., concentrations of virus RNA shed from individuals, proportion of infected individuals shedding, etc.), the model estimated a median range of 171 to 1090 infected persons in the catchment, which agrees with the number of reported cases (Ahmed et al., 2020).
The study from the Netherlands looked for presence/absence of COVID-19 virus RNA fragments. They reported the ability to detect COVID-19 virus RNA with a high level of sensitivity with RT-PCR in the WRRF influent from various WRRFs in the Netherlands. They also report the inability to detect any RNA in the WRRF effluent, although these results are preliminary.
The study by Wu et al. (2020b) quantified several COVID-19 virus genes and estimated the number of virus particles to be approximately 100 gene copies/mL of wastewater, with the lower reported concentration of about 10 gene copies/mL in some samples. Wu et al. (2020b) applied a heat treatment to the wastewater samples to inactivate all viruses for safety reasons. Thus, the concentrations reported by this study are all of non-infectious COVID-19 virus RNA. Unlike the study from Australia, Wu et al. (2020) state that there is a discrepancy between the number of cases calculated by their work on detecting viral RNA in wastewater and the reported number of cases in the population. They highlight the role this approach could play in recommending social distancing or quarantine in communities experiencing infections. In all cases, the studies suggested that detection of the virus RNA in wastewater, even when the COVID-19 prevalence is low, indicates that wastewater surveillance could be used as an early warning tool for increased circulation in the coming winter or unaffected populations.
There are additional ongoing efforts to monitor wastewater for COVID-19 virus RNA, including efforts by the University of Arizona; Stanford University; Michigan State University; Notre Dame University; British Columbia Center for Disease Control in Canada; the Swiss Federal Institute of Technology (EPFL) in Lausanne, Switzerland; France; Spain; Turkey; and the Commonwealth Scientific and Industrial Research Organisation (CSIRO) in Canberra, Australia.
Can COVID-19 virus spread through the fecal-oral contamination route?
Summary: There is currently no evidence of fecal-oral transmission of COVID-19; however, scientific evidence supports the possibility of this transmission route for the following reasons:
- COVID-19 virus was found to bind to receptors in the lungs and intestinal tract; its ability to infect and reproduce in the intestinal tract suggests that it can be excreted with feces in a potentially infective state.
- COVID-19 RNA has been recovered from a large percentage of COVID-positive patients’ fecal material and anal swabs. It also has been recovered from toilet bowls and sinks from positive patients’ rooms.
- Virus RNA was recovered from untreated wastewater in three studies but was not detected in treated wastewater in the one study where it was analyzed.
- Infective virus was detected in feces in two studies without quantification. However, other reports have documented the inability to detect infective virus in stool samples from positive patients.
- Infective virus shows multiple shedding routes, including saliva and respiratory secretions.
- Virus can be highly stable under some conditions, e.g., at room temperatures in laboratory conditions.
Implications for water professionals: Due to the multiple shedding routes of the virus from infected individuals, COVID-19 virus, or at least its RNA, may reach collections systems and WRRFs in wastewater. The virus has the potential to be present in sanitary sewer overflows (SSOs) and combined sewer overflows (CSOs).
While much about COVID-19 virus transmission remains unknown, experts currently believe that exposure to wastewater is not a significant transmission route for COVID-19 virus. The evidence from the Netherlands, however, preliminarily supports the recommendations made by WHO, CDC, OSHA, and WEF and confirms that wastewater treatment can be effective at managing viral risk.
This treatment includes such practices as oxidation with hypochlorite and peracetic acid, as well as inactivation through the use of UV irradiation. We also continue to recommend following regular OSHA recommendations on protective practices for wastewater workers, as shown in Figure 4. As the number of cases continue to increase, it will be important to have a communication plan in place to address any public concerns related to wastewater and drinking water safety. This plan should include how best to address information from sources with weak to no scientific evidence. It is also important to examine on-site operations and practices by maintenance staff and contractors to confirm compliance with WRRF safety and hygiene practices by all (e.g., custodial staff, construction workers).

Figure 4. How to Stay COVID-19 Free at the WRRF. (Get PDF)
Details:
COVID-19 virus in wastewater: At the time of publication of this update, three relevant studies have investigated the use of wastewater-based epidemiology approaches to support public health surveillance for COVID-19 infections in communities. One publication is a pre-proof from Australia by et al.Ahmed et al. (2020), while the other two are pre-prints (i.e., have not been peer-reviewed yet) by Medema et al. (2020) from KWR Watercycle Research Institute in the Netherlands and by Wu et al. (2020b) from Biobot in Massachusetts. Wastewater-based epidemiology relies on using influent COVID-19 virus RNA concentrations in relation to per capita mass loads, daily flows, and size of WRRF population served to estimate the number of infections on a population-scale within sewerage watershed boundaries. All three studies combine RT-PCR with the environmental surveillance approach adopted in the 1940s for monitoring occurrence of polio in wastewater to guide infection control measures.
The work from Australia was the first peer-reviewed publication to confirm the viability of the wastewater-based epidemiology approach for COVID-19 infections. They tested COVID-19 virus RNA concentrations in the wastewater and used these concentrations to estimate the number of infected individuals in the catchment via Monte Carlo simulations. Given the uncertainty and variation in the input parameters (e.g., concentrations of virus RNA shed from individuals, proportion of infected individuals shedding, etc.), the model estimated a median range of 171 to 1090 infected persons in the catchment, which agrees with the number of reported cases (Ahmed et al., 2020).
On March 24, 2020, the KWR Watercycle Research Institute published preliminary results that announce the first detection of COVID-19 virus RNA in untreated wastewater. The study, led by Gertjan Medema, stated that its researchers were unable to detect the virus in treated wastewater, which supports statements made by various agencies (e.g., CDC, OSHA, WHO). The Dutch team collected untreated wastewater samples from WRRFs serving seven cities in the Netherlands on three dates: one before and two during the COVID-19 outbreak. The samples collected prior to the outbreak were all negative and multiple samples from all four WRRFs collected on the last date were positive. The team was only able to analyze treated wastewater from one WRRF but will continue to update the website with new findings.
The team conducted a qualitative risk assessment based on their results to estimate if the risk of COVID-19 infection to employees at WRRF in contact with wastewater or aerosols; the risk assessment reported the risk to be low. Reports are available in Dutch of STOWA and RIONED. The Dutch findings align with the most recent CDC and OSHA assessments, which also describe risk to wastewater workers as low but recommend caution.
All organizations providing guidance on this issue are operating under the assumption that wastewater contains infectious pathogens of concern on a regular basis. We encourage WRRF managers and workers to comply with CDC and OSHA standards, take precautionary measures, and implement all recommended controls for occupational and public health protection.
Wu et al. (2020b) from Biobot quantified the presence of COVID-19 virus RNA at a WRRF in Massachusetts. The team quantified several genes and estimated the number of virus particles to range from approximately 10 to 100 gene copies/mL. They state that there is a discrepancy between the number of cases calculated by their work in detection of virus RNA in wastewater and the reported number of cases in the community, which highlights the role this method could play in wastewater epidemiology. Similar studies are being undertaken globally by various researchers.
COVID-19 virus in the laboratory: A study by Chin et al. (2020) was the first to evaluate the viability of COVID-19 virus in virus transport medium at different temperatures. COVID-19 virus was highly stable at 4°C, displaying an approximate reduction of 0.7 log-units of infectious titre on day 14. It was, however, heat sensitive and completely inactivated at an incubation temperature of 70°C for 5 minutes and highly susceptible to disinfection.
COVID-19 in feces and bodily secretions: While there are limited data on the concentrations of enveloped viruses in feces and urine (Wigginton and Boehm, 2020), various reports have now confirmed the presence of COVID-19 virus RNA in stool samples from positive patients (Wu et al., 2020; Chen et al., 2020; Holshue et al., 2020; Xiao et al., 2020; Ong et al., 2020; Tang et al., 2020b; Wölfel et al., 2020). Virus shedding patterns in feces are highly variable and present some challenges for modeling the number of infections in the community. Wang et al. (2020) reported the percentage of RNA positive fecal samples in a sample of 205 patients to be 29%. Chen et al. (2020) reported 64% of fecal samples to be RNA positive in a sample of 44 patients. Wölfel et al. (2020) reported that stool samples remained RNA-positive over three weeks in six of the nine examined patients, in spite of full resolution of symptoms. Wölfel et al. (2020) also remarked that no infective virus was detected in spite of high RNA concentrations in feces. The authors recommended that future work evaluate if exposure to the gut environment inactivates COVID-19 virus.
To this date, only two studies (Zhang et al., 2020a; Wang et al., 2020) were able to detect infective virus in feces. These studies confirmed viral persistence with electron microscopy but did not quantify the viral particles detected. The latter suggested that the detection of infective virus is highly dependent on virus concentrations in feces (Wang et al., 2020). It is difficult to estimate the number of studies unable to isolate infectious virus in feces since scientific studies do not usually publish negative results (i.e., when the virus is not detected) as it may only be a reflection of deficiencies in sample collection, preparation, or analysis. For example, recent work has noted that, unlike SARS-CoV and MERS-CoV, infected patients showing symptoms of gastrointestinal infection at later stages of infection, COVID-19 patients may harbour the virus in the intestine at the early or late stage of disease (Zhang et al., 2020b). Wu et al (2020a) suggested that the COVID-19 virus RNA can persist in feces for nearly 5 weeks even when the patients' respiratory samples tested negative for the virus. Also, Tang et al (2020b) detected virus RNA in stool specimens from an asymptomatic child for at least 9 days, where respiratory tract specimens were negative. Since COVID-19 virus seems to defy many of our assumptions about disease progression, it may be important for researchers to examine fecal material and anal swabs for viable COVID-19 virus and RNA early on during infection as well as in later stages.
It is also noteworthy that infective virus was isolated from saliva (To et al., 2020) and various respiratory secretions (Peng et al., 2020; Zhang et al., 2020b) that are commonly released into the collection system in domestic wastewater. This implies that COVID-19 virus may reach the WRRF through bodily secretions other than feces from infected individuals.
COVID-19 has host cells in the respiratory and intestinal tract: Additionally, recent work has shown that COVID-19 virus binds to the angiotensin-converting enzyme 2 (ACE2) receptors on Type II alveolar cells and intestinal epithelia (Hoffman et al., 2020; Wan et al., 2020; Wrapp et al., 2020). Its ability to attach to these cell receptors in the intestinal tract supports the potential for fecal-oral transmission.
Occupational transmission of COVID-19 at WRRF: The viability of COVID-19 virus under environmental conditions that may facilitate its transmission via the fecal–oral pathway is not yet clear (Wigginton and Boehm, 2020). Despite the lack of epidemiological evidence, various authors have cautioned in editorial pieces that this may present a problem in conditions where wastewater treatment, sanitation, and hygiene are challenged (Lodder and De Roda Husman, 2020; Yeo et al., 2020). In the developed world, it is important to note that no infections due to occupational exposure have been reported to date.
Can COVID-19 virus be transmitted via aerosols?
Summary: Limited, but contradictory, evidence supports the potential survival of infective COVID-19 virus in aerosols. Various studies confirm that control measures (e.g., ventilation, no crowds, open spaces, proper disinfection) can effectively limit transmission. Experts also highlight that plumbing systems in buildings need to be monitored with caution.
Implication for water professionals: While infective virus has not yet been isolated from wastewater, we should take a precautionary approach and assume that COVID-19 virus, like other common viral and bacterial pathogens found in wastewater, could survive in aerosols when wastewater is mechanically agitated, mixed, or aerated. Good ventilation, limited crowding, and other precautionary measures are effective at reducing virus concentrations in aerosols. We encourage WRRF managers and workers to take precautionary measures and implement all recommended controls for occupational and public health protection.
WEF has identified a water sector need for additional guidance on bioaerosols exposure and protective measures at WRRFs and has convened a Blue Ribbon Panel to examine these issues.
Details: While droplet transmission (>5 μm) is thought to be the major mode of transmission for COVID-19 virus, data on survival in aerosols (<5 μm) has been limited so far. It is important to recognize that this may be related to the difficulty in sampling and characterizing the viability of airborne viruses (Morawski and Cao, 2020), which is highly dependent on the virus type, environmental conditions, and on the methods of collection and handling of bioaerosol samples (Cao et al., 2011).
One study led by the NIH confirmed infective COVID-19 virus survival for more than 3 hours in experimentally generated aerosols under laboratory conditions. The study concluded that the half-lives of COVID-19 virus and SARS-CoV-1 in air were comparable at 1.1 to 1.2 hours under the conditions tested. They proposed that differences in resulting disease epidemiology are not related to differences in environmental survival but to other characteristics of COVID-19 virus (e.g., high load in upper respiratory tract, asymptomatic transmission). More recently, a pre-print by Fears et al. (2020) supported these findings confirming that in laboratory conditions infectious COVID-19 virus was detected at all timepoints during the aerosol suspension stability experiment for up to 16 hours.
Outside the laboratory setting, however, the results are contradictory. An article in Nature suggests that COVID-19 virus RNA concentrations were very low and declined significantly in aerosol samples from two hospitals in Wuhan after patient numbers were reduced post-outbreak and rigorous disinfection measures were implemented. The same study highlighted that the majority of aerosol samples from public spaces were negative with crowding impacting the results. Another study that investigated aerosols in three COVID patients' rooms was unable to recover any COVID-19 virus from air. Finally, an epidemiological investigation by Cai et al. (2020) into a COVID-19 cluster of cases at a mall in Wenzhou, China, was unable to find a direct transmission cause and suggested that indirect transmission by aerosols in a confined space (restroom or elevator) may have occurred. However, these studies could not rule out possible exposure and transmission from contact surfaces such as handrails, doorknobs, etc. The last two studies highlight that control measures, such as ventilation and open space and proper use and standard disinfection of bathroom surfaces, can effectively limit aerosol transmission of COVID-19 virus.
A recent publication in the Lancet (Gormley et al., 2020) has cautioned that the risk of transmission of COVID-19 virus in building plumbing systems should not be underestimated. In those circumstances, the fresh fecal material is more likely to contain infective virus and conditions for aerosolization exist. The authors drew on the SARS outbreak from Amoy Gardens in Hong Kong and outlined a set of recommendations, including making sure that all water appliances in bathrooms and kitchens are fitted with a functioning U-bend, preventing the loss of the water trap seal within a U-bend, and opening a tap on all water appliances for at least 5 seconds twice a day (morning and evening) paying special attention to floor drains in bathrooms.
Evidence of COVID-19 virus survival on surfaces
Summary: The available evidence, although limited, supports survival of COVID-19 virus on surfaces and the efficacy of disinfectants at reducing transmission.
Implication for water professionals: Limited evidence suggests that COVID-19 virus, like other common waterborne viral and bacterial pathogens, can survive on surfaces for up to 9 days. Disinfecting PPE, frequently touched surfaces, surfaces where aerosols may be deposited or surfaces that come into contact with wastewater from splashing is recommended. Evidence suggests that disinfecting inanimate surfaces with 0.1% to 0.5% sodium hypochlorite, 0.5% peracetic acid, or 70% ethanol for 1 minute or longer should be effective.
Details: Several relevant reports have been published on COVID-19 virus survival on surfaces and efficacy of surface disinfection practices. Kampf et al. (2020) conducted a meta-analysis reviewing 22 published papers evaluating the survival of human and veterinary coronaviruses on inanimate surfaces. They report that coronaviruses can survive on surfaces up to nine days and that good disinfection practices can be protective. They recommend a dilution of 1:50 of 5% sodium hypochlorite (resulting in a final dilution of 0.1%) or 0.5% hydrogen peroxide, and a contact time of 1 minute for larger areas. For smaller areas, they recommend 70% ethanol (62% to 71%; carrier tests) as per WHO recommendations. They report that some biocides (e.g., 0.05% to 0.2% benzalkonium chloride or 0.02% chlorhexidine digluconate) tend to be somewhat less effective.
Other studies looking at COVID-19 virus viability specifically in laboratory conditions, as opposed to surrogates, corresponded with the results of this meta-analysis. van Doremalen (2020) documented the detection and exponential decay of infective virus on plastic, stainless steel, copper, and cardboard reporting their half-lives to be 6.8, 5.6, 0.7 and 3.5 hours, respectively. Similarly, Chin et al. (2020) confirmed that SARS-CoV-2 was stable and infective on smooth surfaces compared to printing and tissue paper, surviving up to 4 days on glass and bank notes and 7 days on stainless steel and plastic. The same study showed that eight commonly used disinfectants diluted to recommended concentrations all inactivated COVID-19 virus after 5 minutes of contact time. The least effective of these disinfectants were soap and water (1:49 dilution), which still achieved a 3.6 log reduction in virus titres after a 5-minute exposure time.
The remaining studies relied on virus RNA for detection. Ong et al. (2020) tested fomites in three COVID-19 patients’ rooms and found virus RNA in toilet bowls and sink samples, as well as on PPE — specifically, shoe covers. The team reported that post-cleaning samples were all negative, suggesting that current decontamination measures are sufficient. Another study examined viral RNA in three types of aerosol samples: total suspended particle, size segregated, and deposition aerosol. Their early results support the possibility of virus aerosol deposition on PPE or floor surfaces and their subsequent resuspension as a potential transmission pathway. Finally, Cai et al. (2020) reported the possible occurrence of indirect transmission in Wenzhou (China). The Guangzhou Center for Disease Control and Prevention detected the COVID-19 virus RNA on a doorknob in a patient’s house, but were unable to find COVID-19 virus on any of the examined mall surfaces (e.g., mall elevator walls and buttons).
All of the studies cited confirm the importance of surface disinfection and protective measures. This is particularly important in the context of the proposed low infective dose of COVID-19 virus that some experts suspect.
Comparing SARS, MERS and COVID-19 viruses
Summary: COVID-19 virus has been shown to be more transmissible than SARS-CoV and MERS-CoV. Like SARS-CoV, it uses ACE2 receptors on host cells in the respiratory and gastrointestinal systems to invade host cells and can survive in aerosols and fomites for hours.
Implications for water professionals: Despite differences in resulting illness, data extrapolated from SARS-CoV and MERS-CoV (as well as other surrogates) have been helpful in predicting COVID-19 virus behaviour and recommending appropriate risk management practices before analyses specific to COVID-19 virus is completed and validated.
Details: Compared with the known SARS-CoV and MERS-CoV genome, COVID-19 virus is closer to the SARS-like bat CoVs in terms of the whole genome sequence. Most genomic encoded proteins of COVID-19 virus are similar to SARS-CoV, though certain differences do exist. Several independent research groups have identified that COVID-19 virus belongs to β-coronavirus group, with high genomic similarity to bat coronavirus. Though this points to bats as the natural host, the identity of an intermediate host is still not confirmed.
Current reports still show that COVID-19 virus may be more transmissible, while causing less serious illness or virulence compared to SARS-CoV and MERS-CoV (Guo et al., 2020). As of March 1, 2020, the currently analyzed mortality of COVID-19 is 3.4%, lower than death rate of SARS (9.6%) and MERS (around 35%), respectively (de Wit et al., 2016).
Binding to the host cells is a critical step for viral cell entry and infection. Studies have shown that the COVID-19 virus binds to host cells through its spike glycoprotein and uses the same host cell receptors — angiotensin-converting enzyme 2 (ACE2) – as that for SARS-CoV tract (Hoffman et al., 2020; Wan et al., 2020). ACE2 protein is present in abundance on lung alveolar epithelial cells and the enterocytes of small intestine, which may explain the routes of infection and disease manifestations (Guo et al., 2020). A study by Wrapp et al. (2020), compared the biophysical strength of binding between the glycoprotein spikes of the COVID-19 virus and the SARS-CoV and found this protein binds at least ten times more tightly than the corresponding spike protein of SARS–CoV to their common host cell receptor. The authors proposed that this may explain the higher transmission rate of COVID-19 from human to human; however, additional studies are needed to investigate this possibility. MERS-CoV, on the other hand, uses dipeptidyl peptidase 4 (also known as CD26).
Comparisons of aerosol and fomite survival of SARS-CoV and COVID-19 virus have shown that the viruses have very similar half-lives and that differences in disease transmission stem from other infection characteristics.
Authors
- Rasha Maal-Bared is the Senior Microbiologist at EPCOR Water Services Inc. (Edmonton, Canada) and the current chair of the Waterborne Infection Disease Outbreak Control Working Group.
- Naoko Munakata is a Supervising Engineer at the Los Angeles County Sanitation Districts.
- Kyle Bibby is an Associate Professor and the Wanzek Collegiate Chair in the Department of Civil and Environmental Engineering and Earth Sciences at the University of Notre Dame (Notre Dame, Ind.).
- Kari Brisolara is the Associate Dean for Academic Affairs and an Associate Professor of Environmental and Occupational Health Sciences at the Louisiana State University Health Sciences Center (New Orleans, La.) and Vice Chair of the WEF Disinfection and Public Health Committee.
- Charles Gerba is a professor of epidemiology and biostatistics in the Dept of Environmental Science at the University of Arizona and a supporting member of the WIDOC working group.
- Mark Sobsey was the Kenan Distinguished Professor of Environmental Sciences and Engineering at the Gillings School of Global Public Health at the University of North Carolina (Chapel Hill). He has served as an advisor on numerous national and international drinking water and WASH committees for the World Health Organization, WEF, and AWWA.
- Scott Schaefer is the Wastewater Practice Leader with AE2S (Saint Joseph, Minn.) and the Chair of the WEF Disinfection and Public Health Committee.
- Lee Gary is an Adjunct Professor at Tulane University, an instructor with the Basic Academy at the FEMA/Emergency Management Institute (Emmitsburg, Md.) and the owner and CEO of Strategic Management Services (New Orleans).
- Samendra Sherchan is an Assistant Professor at the Tulane School of Public Health and Tropical Medicine (New Orleans, La.).
- Jay Swift is a Principal Engineer with Gray and Osborne (Seattle, Wash.).
- Akin Babatola is the Laboratory and Environmental Compliance Manager at the City of Santa Cruz (San Francisco Bay Area).
- Lola Olabode is a Program Director at the Water Research Foundation and an expert in risk management during outbreaks (Washington, D.C.).
- Robert S. Reimers is a Professor Emeritus at Tulane University’s School of Public Health and Tropical Medicine, the Director of Asepticys Inc. (New Orleans, La.).
- Robert Bastian is a 2016 WEF Fellow and water expert.
- Albert Rubin is a professor emeritus at North Carolina State University in the Dept of Biological and Agricultural Engineering.
Acknowledgements: The authors would like to thank Dr. Charles Haas (Drexel University, Philadelphia) for reviewing the content of this update and its associated conclusions.
Disclaimers: The information reported here is a summary of current knowledge about this emerging viral pathogen. The state of knowledge will evolve as additional investigation and research is conducted, so continuous review of reputable sources and websites is advised.
The conclusions, findings, and opinions expressed by the authors contributing to this piece do not reflect the official position of the U.S. Occupational Safety and Health Administration, the U.S. Centers for Disease Control and Prevention, the U.S. Environmental Protection Agency, or any authors' affiliated institution.
•••
Monitor WEF’s coronavirus resource page for the most up-to-date information and resources for the water sector.
Download a PDF of this article:
Coronavirus and Water
(WSEC-2020-TR-002)
•••
Notable Updates:
Since the date of publication of this article, notable new and updated research has emerged. You can view these notable updates here.
•••
Relevant References
Ahmed, W., Angel, N., Edson, J., Bibby, K., Bivins, A., O'Brien, J. W., ... & Tscharke, B. (2020). First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Science of The Total Environment, 138764. https://www.sciencedirect.com/science/article/pii/ S0048969720322816
Cai, J., Sun, W., Huang, J., Gamber, M., Wu, J., & He, G. (2020). Indirect Virus Transmission in Cluster of COVID-19 Cases, Wenzhou, China, 2020. Emerging infectious diseases, 26(6). https://wwwnc.cdc.gov/eid/article/26/6/20-0412_article
Cao, G., Noti, J. D., Blachere, F. M., Lindsley, W. G., & Beezhold, D. H. (2011). “Development of an improved methodology to detect infectious airborne influenza virus using the NIOSH bioaerosol sampler.” Journal of Environmental Monitoring, 13(12), 3321-3328.
Chen, Y., Chen, L., Deng, Q., Zhang, G., Wu, K., Ni, L., ... & Yang, J. (2020). “The Presence of SARS‐CoV‐2 RNA in Feces of COVID‐19 Patients.” Journal of Medical Virology. https://onlinelibrary.wiley.com/doi/full/10.1002/jmv.25825
Chin, A., Chu, J., Perera, M., Hui, K., Yen, H. L., Chan, M., ... & Poon, L. (2020). “Stability of SARS-CoV-2 in different environmental conditions.” The Lancet April 2020: https://www.thelancet.com/journals/lanmic/article/PIIS2666-5247(20)30003-3/fulltext
de Wit, E., van Doremalen, N., Falzarano, D., & Munster, V. J. (2016). “SARS and MERS: recent insights into emerging coronaviruses.” Nature Reviews Microbiology, 14(8), 523.
Fears, A. C., Klimstra, W. B., Duprex, P., Hartman, A., Weaver, S. C., Plante, K. S., ... & Nalca, A. (2020). “Comparative dynamic aerosol efficiencies of three emergent coronaviruses and the unusual persistence of SARS-CoV-2 in aerosol suspensions.” medRxiv. https://www.medrxiv.org/content /10.1101/ 2020.04.13.20063784v1
Gormley, M., Aspray, T. J., & Kelly, D. A. (2020). ”COVID-19: mitigating transmission via wastewater plumbing systems.” The Lancet Global Health.
Guo, Y. R., Cao, Q. D., Hong, Z. S., Tan, Y. Y., Chen, S. D., Jin, H. J., ... & Yan, Y. (2020). “The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak–an update on the status.” Military Medical Research, 7(1), 1-10.
Hoffmann, M., Kleine-Weber, H., Krüger, N., Mueller, M. A., Drosten, C., & Pöhlmann, S. (2020). “The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells.” BioRxiv. https://www.biorxiv.org/content/10.1101/ 2020.01.31.929042v1.abstract
Holshue, M.L., et al. First Case of 2019 Novel Coronavirus in the United States. New England Journal of Medicine (2020). DOI: 10.1056/NEJMoa2001191.
Kampf, G., Todt, D., Pfaender, S., & Steinmann, E. (2020). “Persistence of coronaviruses on inanimate surfaces and its inactivation with biocidal agents.” Journal of Hospital Infection.
Lee, P. I., & Hsueh, P. R. (2020). “Emerging threats from zoonotic coronaviruses-from SARS and MERS to 2019-nCoV.” Journal of Microbiology, Immunology and Infection. https://www.sciencedirect.com/science/article/pii/ S1684118220300116?via%3Dihub
Liu, Y., Ning, Z., Chen, Y. et al. ”Aerodynamic analysis of SARS-CoV-2 in two Wuhan hospitals.” Nature (2020). https://doi.org/10.1038/s41586-020-2271-3. https://www.nature.com/articles/s41586-020-2271-3#citeas
Lodder, W., de Roda Husman A.M. (2020). “SARS-CoV-2 in wastewater: potential health risk, but also data source.” The Lancet Gastroenterology and Hepatology. DOI: https://doi.org/10.1016/S2468-1253(20)30087-X
Mallapati, S. 2020. “How sewage can reveal true scale of coronavirus outbreak.” Nature April 3, 2020. https://www.nature.com/articles/d41586-020-00973-x#correction-0
Medema, G., Heijnen, L., Elsinga, G., Italiaander, R., Brouwer, A. (2020) “Presence of SARS-Coronavirus-2 in sewage.” MedRxiv. Preprint DOI: https://doi.org/10.1101/2020.03.29.20045880
Metcalf, T. G., Melnick, J. L., & Estes, M. K. (1995). “Environmental virology: from detection of virus in sewage and water by isolation to identification by molecular biology—a trip of over 50 years.” Annual review of microbiology, 49(1), 461-487.
Morato, J., Mir, J., Codony, F., Mas, J. & Ribas F. (2003) “Microbial response to disinfectants.” Mara, D., & Horan, N. J. (Eds.). (2003). Handbook of water and wastewater microbiology. Elsevier, pp. 658-693.
Morawska, L., Cao, J. (2020). “Airborne transmission of SARS-COV-2: the world should face the reality.” Environment International 139: 105730.
Munster, V. J., Koopmans, M., van Doremalen, N., van Riel, D., & de Wit, E. (2020). “A novel coronavirus emerging in China—key questions for impact assessment.” New England Journal of Medicine, 382(8), 692-694. https://www.nejm.org/doi/full/10.1056/NEJMp2000929
Ong, S. et al. (2020). “Air, Surface Environmental, and Personal Protective Equipment Contamination by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) From a Symptomatic Patient.” JAMA.
OSHA/CDC Guidance: https://www.osha.gov/SLTC/covid-19/controlprevention.html#solidwaste
Peng, X., Xu, X., Li, Y., Cheng, L., Zhou, X., & Ren, B. (2020). “Transmission routes of 2019-nCoV and controls in dental practice.” International Journal of Oral Science, 12(1), 1-6.
Prussin, A. J., Belser, J. A., Bischoff, W., Kelley, S. T., Lin, K., Lindsley, W. G., ... & Marr, L. C. (2020). Viruses in the Built Environment (VIBE) meeting report.
Rodriguez-Lazaro, D., Cook, N., Ruggeri, F. M., Sellwood, J., Nasser, A., Nascimento, M. S. J., ... & Bosch, A. (2012). “Virus hazards from food, water and other contaminated environments.” FEMS microbiology reviews, 36(4), 786-814.
Tang, X., Wu, C., Li, X., Song, Y., Yao, X., Wu, X., ... & Cui, J. (2020a). “On the origin and continuing evolution of SARS-CoV-2.” National Science Review. https://academic.oup.com/nsr/advance-article/doi/10.1093/nsr/nwaa036/5775463
Tang, A., Tong, Z. D., Wang, H. L., Dai, Y. X., Li, K. F., Liu, J. N., ... & Yan, J. B. (2020b). “Detection of Novel Coronavirus by RT-PCR in Stool Specimen from Asymptomatic Child, China.” Emerging infectious diseases, 26(6).
Thomas, Y., Vogel, G., Wunderli, W., Suter, P., Witschi, M., Koch, D., ... & Kaiser, L. (2008). “Survival of influenza virus on banknotes.” Appl. Environ. Microbiol., 74(10), 3002-3007.
To, K. K. W., Tsang, O. T. Y., Chik-Yan Yip, C., Chan, K. H., Wu, T. C., Chan, J., ... & Lung, D. C. (2020). “Consistent detection of 2019 novel coronavirus in saliva.” Clinical Infectious Diseases.
Torrey, J., von Gunten, U., & Kohn, T. (2019). “Differences in viral disinfection mechanisms as revealed by quantitative transfection of Echovirus 11 genomes.” Applied and environmental microbiology, 85(14), e00961-19.
van Doremalen, N., Bushmaker, T., Morris, D. H., Holbrook, M. G., Gamble, A., Williamson, B. N., ... & Lloyd-Smith, J. O. (2020). “Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1.” New England Journal of Medicine.
Wan, Y., Shang, J., Graham, R., Baric, R. S., & Li, F. (2020). “Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus.” Journal of Virology, 94(7). https://jvi.asm.org/content/94/7/e00127-20.abstract
Wang et al., (2005a). “Study on the resistance of severe acute respiratory syndrome associated coronviruses.” Journal of Virological Methods 126(1-2), 171-177.
Wang, Xin-Wei, et al. (2005b) “Concentration and detection of SARS Coronavirus in sewage from Xiao Tang Shan Hospital and the 309th Hospital.” Journal of Virological Methods 128.1-2: 156-161.
Wang, W., Xu, Y., Gao, R., Lu, R., Han, K., Wu, G., & Tan, W. (2020). “Detection of SARS-CoV-2 in Different Types of Clinical Specimens.” JAMA. https://jamanetwork.com/journals/jama/fullarticle/2762997
Water Environment Federation. 2020. The Water Professional's Guide to COVID-19.
Wigginton, K. R., Pecson, B. M., Sigstam, T., Bosshard, F., & Kohn, T. (2012). “Virus inactivation mechanisms: Impact of disinfectants on virus function and structural integrity.” Environmental science & technology, 46(21), 12069-12078.
Wigginton, K. R., & Boehm, A. B. (2020). "Environmental Engineers and Scientists Have Important Roles to Play in Stemming Outbreaks and Pandemics Caused by Enveloped Viruses.” Environment Science & Technology https://pubs.acs.org/doi/full/10.1021/acs.est.0c01476
Wölfel, R., Corman, V.M., Guggemos, W. et al. ”Virological assessment of hospitalized patients with COVID-2019.” Nature (2020). https://doi.org/10.1038/s41586-020-2196-x
World Health Organization. 2020. Water, Sanitation, Hygiene and Waste Management for COVID-19.
Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C. L., Abiona, O., ... & McLellan, J. S. (2020). “Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation.” Science, 367(6483), 1260-1263.
Wu, Y., Guo, C., Tang, L., Hong, Z., Zhou, J., Dong, X., ... & Kuang, L. (2020a). “Prolonged presence of SARS-CoV-2 viral RNA in faecal samples.” The Lancet Gastroenterology & Hepatology.
Wu, F.Q., Xiao, A., Zhang, J.B., Gu, X.Q., Lee W.L., Kauffman, K… & Alm, E.J. (2020b). “SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases.” medRxiv. https://www.medrxiv.org/content/10.1101/ 2020.04.05.20051540v1.full.pdf
Xiao, F., et al. (2020). “Evidence for gastrointestinal infection of SARS-CoV-2.” Gastroenterology March 2020. DOI: https://doi.org/10.1053/j.gastro.2020.02.055
Yeo, C., Kaushal, S., & Yeo, D. (2020). “Enteric involvement of coronaviruses: is faecal–oral transmission of SARS-CoV-2 possible?.” The Lancet Gastroenterology & Hepatology, 5(4), 335-337. https://www.thelancet.com/journals/langas/article/PIIS2468-1253(20)30048-0/fulltext
Yuen, K. S., Ye, Z. W., Fung, S. Y., Chan, C. P., & Jin, D. Y. (2020). “SARS-CoV-2 and COVID-19: The most important research questions.” Cell & bioscience, 10(1), 1-5.
Zhang, Y. et al. 2020a. “Isolation of 2019-nCoV from a Stool Specimen of a Laboratory-Confirmed Case of the Coronavirus Disease 2019 (COVID-19)[J].” China CDC Weekly, 2020b, 2(8): 123-124.
Zhang, W. et al. (2020b) “Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes.” Emerging Microbes & Infections, 9:1, 386-389. DOI: 10.1080/22221751.2020.1729071
Glossary
Airborne transmission: Can occur in two ways: either through relatively large particles of respiratory fluid called droplets (101 to 102 μm diameter) or through smaller particles called aerosols that can remain aerosolized for longer periods of time called droplet nuclei (<101 μm diameter). As larger droplets are pulled to the ground by gravity quickly, droplet transmission requires close physical proximity between infected and susceptible individuals, whereas aerosolized transmission can occur over larger distances and does not necessarily require that infected and susceptible individuals are at the same location at the same time. Diseases like measles and chickenpox result in the production of infective bioaerosols for hours, while COVID-19 is mainly transmitted by respiratory droplet.
Aerosols: Refers to situations where droplet nuclei (residue from evaporated droplets) or dust particles containing microorganisms can remain suspended in air for long periods of time. These organisms must be capable of surviving for long periods of time outside the body and must be resistant to drying. Airborne transmission allows organisms to enter the upper and lower respiratory tracts. Fortunately, only a limited number of diseases are capable of airborne transmission, such as measles, chickenpox, and tuberculosis.
Asymptomatic: Situation when a person is carrying a disease but is not showing signs of infection. In COVID-19 infections, asymptomatic transmission has been reported.
Capsid: A protein coat that encapsulates a virus’ genetic material.
Cell culture: Bacteria can be grown on artificial nutrient media in the presence of appropriate environmental conditions. Viruses, however, require living host cells for replication. In cell culture, the sample is introduced to a “culture” of appropriate host cells (eukaryotic or prokaryotic) in an artificial environment (test tube, culture flask, agar plate) and effects on host cells can be monitored.
Direct transmission: Requires close physical contact (touching, contact with oral secretions, etc.) between an infected person and a susceptible person, and the physical transfer of microorganisms. It will usually occur between members of the same household or close friends and family. Diseases spread exclusively by direct contact are unable to survive for significant periods of time away from a host.
DNA: Deoxyribonucleic acid is a molecule composed of two polynucleotide chains that coil around each other to form a double helix carrying genetic instructions for the development, functioning, growth, and reproduction of all known organisms and many viruses. DNA and ribonucleic acid (RNA) are nucleic acids.
Droplet contact: Primary cause of COVID-19 and SARS infections. Refers to transmission of diseases by infected droplets contacting surfaces of the eye, nose, or mouth, where the droplets containing infectious agents are generated when an infected person coughs, sneezes, or talks. Droplets can also be generated during certain medical procedures, such as bronchoscopy. Droplet transmission can be reduced with the use of personal protective barriers, such as face masks and goggles.
Fecal-oral transmission: Fecal-oral transmission is usually associated with organisms that infect the digestive system. Microorganisms enter the body through ingestion of contaminated food and water. Inside the digestive system (usually within the intestines) these microorganisms multiply and are shed from the body in feces. If proper hygienic and sanitation practices are not in place, the microorganisms in the feces may contaminate the water supply through inadequate wastewater treatment or enter the body through inadequate hygiene practices and hand washing.
Fomites: Fomites consist of both porous and nonporous surfaces or objects that can become contaminated with a pathogen and serve as vehicles in transmission of infection. Fomite exposure requires an inanimate object to carry a pathogen from one susceptible animal to another. Fomite exposure often involves a secondary route of exposure such as oral or direct contact for the pathogen to enter the host.
Genome: The genetic material of an organism.
Inactivation: Rendering infectious agents unable to infect cells through physical, chemical, or biological means thereby removing health risk through exposure. Inactivation is a process that is often combined with physical removal in water and wastewater treatment to increase log reductions in protozoa or viruses.
Indirect transmission: Refers to situations where a susceptible person is infected from contact with a contaminated surface. This is highly reliant on the organism’s persistence (or survival) in the environment.
Infective dose: The amount of virus particles needed to result in infection in a host. Viral infective doses are usually much lower than bacterial infective doses.
Persistence: Similar to survival. Refers to the ability to maintain virus infectivity outside a host in the presence of environmental stressors.
RNA: Ribonucleic acid (RNA) is a molecule essential in various biological roles in coding, decoding, regulation, and expression of genes. Like DNA, RNA is assembled as a chain of nucleotides but is found in nature as a single strand folded onto itself, rather than a paired double strand. Many viruses encode their genetic information using an RNA genome.
Shedding: Release of virus particles from an infected person’s body.
Transfection: A method used in virology to determine if purified nucleic acids remain infectious. It involves introducing the purified genetic material into an animal cells and monitoring the potential development of mature viral particles.
Viable virus: Intact virus outside a host still able to result in infection. Often called “infective” or “live” in scientific literature.
Virion: A virus outside a host cell (also called extracellular virus) in the environment that is infectious. To remain infectious or viable, a virion should still have all its parts intact: an RNA or DNA core with a protein coat and, sometimes, with external envelopes.
Virus: A submicroscopic infectious agent that replicates only inside the living cells of an organism. Viruses are microscopic parasites, generally much smaller than bacteria. With a diameter of 220 nanometers, the measles virus is about 8 times smaller than E. coli bacteria. At 45 nm, the hepatitis virus is about 40 times smaller than E. coli. The polio virus, 30 nm across, is about 10,000 times smaller than a grain of salt. When not inside an infected cell or in the process of infecting a cell, viruses exist in the form of independent particles, or virions, consisting of genetic material a protein coat called a capsid and in some cases a lipid envelope.
When outside a host body, they lack the capacity to thrive and reproduce and are considered to be inert. This means that the virus is completely dependent on the host’s cellular machinery to produce copies of themselves and to build proteins based on the instructions encoded in their RNA.


